This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
bulk shift effect or binding with rapid kinetics
- spring
- Topic Author
- Offline
- New Member
-

Less
More
- Posts: 2
- Thank you received: 0
3 months 2 days ago #1
by spring
bulk shift effect or binding with rapid kinetics was created by spring
Hello,
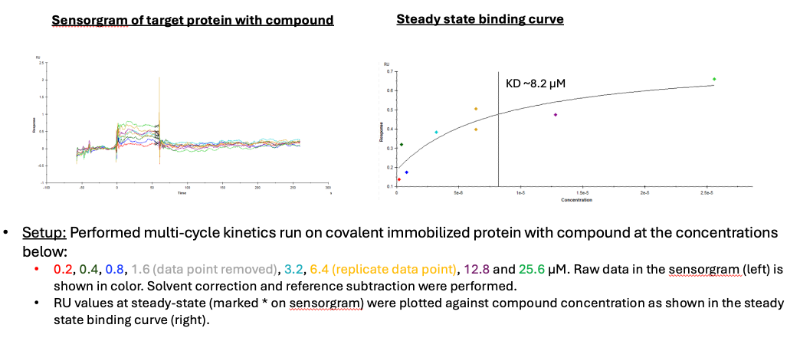
I am a newbie to SPR and would appreciate some advice on how to interpret our sensorgram of a small molecule interaction with our immobilized target protein. I immobilized our protein target (~27 kD) onto a CM5 chip through amine coupling. The amount of immobilized protein is ~1450 RU, which gives a calculated R max of ~25. We do not have prior knowledge of the KD of the compound:protein, nor if the compound does bind. The sensorgram attached shows the multi-cycle kinetic run with compound at a concentration of 25 µM diluted two-fold to 0.2 µM.
The amount of DMSO in the running buffer is 2% and the compound was diluted to 2% DMSO in running buffer without DMSO, then serially diluted 1:1 in running buffer with 2% DMSO. DMSO solvent correction and reference subtraction (Flow cell surface activated and deactivated) were also performed.
In the sensorgram result, we see very boxy response traces for all compound concentrations and the signal is very low. I am not sure if this indicative of just bulk solvent effects, or if there are true binding events occurring with very fast on and off rates, or even a combination of both. The program used the steady-state to generate the binding curve and determined a KD.
Any throughts/suggestions/recommendations on this result?
Thanks you very much for your help in this and please let me know if you need any other info.
I am a newbie to SPR and would appreciate some advice on how to interpret our sensorgram of a small molecule interaction with our immobilized target protein. I immobilized our protein target (~27 kD) onto a CM5 chip through amine coupling. The amount of immobilized protein is ~1450 RU, which gives a calculated R max of ~25. We do not have prior knowledge of the KD of the compound:protein, nor if the compound does bind. The sensorgram attached shows the multi-cycle kinetic run with compound at a concentration of 25 µM diluted two-fold to 0.2 µM.
The amount of DMSO in the running buffer is 2% and the compound was diluted to 2% DMSO in running buffer without DMSO, then serially diluted 1:1 in running buffer with 2% DMSO. DMSO solvent correction and reference subtraction (Flow cell surface activated and deactivated) were also performed.
In the sensorgram result, we see very boxy response traces for all compound concentrations and the signal is very low. I am not sure if this indicative of just bulk solvent effects, or if there are true binding events occurring with very fast on and off rates, or even a combination of both. The program used the steady-state to generate the binding curve and determined a KD.
Any throughts/suggestions/recommendations on this result?
Thanks you very much for your help in this and please let me know if you need any other info.
Please Log in or Create an account to join the conversation.
- Arnoud
-

- Offline
- Moderator
-

Less
More
- Posts: 47
- Thank you received: 6
3 months 1 day ago #2
by Arnoud
Replied by Arnoud on topic bulk shift effect or binding with rapid kinetics
Hi,
You have made it easy to your self. But without seeing all the steps (raw data), I think you are on the right way.
If possible, i would make a second active channel with higher ligand concentration to confirm the interaction is real. It should give more response, proportional with the previous experiment response. Also repeat the experiment and see if you get the same answer.
In addition, you must at least have another type of experiment to confirm the binding like FRET or an inhibition assay.
Kind regards
Arnoud
You have made it easy to your self. But without seeing all the steps (raw data), I think you are on the right way.
If possible, i would make a second active channel with higher ligand concentration to confirm the interaction is real. It should give more response, proportional with the previous experiment response. Also repeat the experiment and see if you get the same answer.
In addition, you must at least have another type of experiment to confirm the binding like FRET or an inhibition assay.
Kind regards
Arnoud
The following user(s) said Thank You: spring
Please Log in or Create an account to join the conversation.
- Sutkeviciute
- Offline
- New Member
-

Less
More
- Posts: 3
- Thank you received: 0
2 months 1 week ago #3
by Sutkeviciute
Replied by Sutkeviciute on topic bulk shift effect or binding with rapid kinetics
Hi! I would include double-referencing and a reference surface with an inactive protein. I am usually doing DMSO "titration" to prepare samples for double-referencing: I would just prepare the DMSO samples identically as a compound, except using DMSO (instead of the compound) as a starting solution. It looks in your case what you are getting just bulk responses, and those 2 adjustments can help you understand if it's true.
Please Log in or Create an account to join the conversation.
Moderators: Arnoud