The ligand
Choosing the correct ligand can have a great influence on the interaction system. For instance: antibodies, like IgG, have two identical binding sites. Using antibodies as an analyte, will give rise to a bivalent analyte model. In principle, this model is useful, assuming that the ligand density is not too low and the dextran matrix is flexible enough. Nevertheless, it is better to avoid this kind of multiple binding site situations by immobilizing the antibody. By using the multivalent protein as ligand the interaction system treats every ligand binding site as a single entity.
However, there are situations where dimerization or multivalence binding is beneficial (1). Receptors often have to dimerize before they activate a signal (2),(3),(4). By immobilizing the single binding site receptors and using a multivalent analyte, multimerization can occur.
Ligand heterogeneity
It is important that the ligand is pure and does not contain other forms. This is easily checked by electrophoresis. Care must be taken though, to ensure that the protein is properly handled and remains active. Ligand heterogeneity may give rise to non-Langmuirian kinetics, which is more difficult to solve.
Also, make sure that heterogeneity caused by immobilization is minimized. Amine coupling results in random immobilization, with different orientations of the ligand. To overcome ligand heterogeneity other immobilization chemistries or ligand capturing strategies can be used.
Ligand density
For most applications, it is important to use the lowest possible ligand density, as this will provide good quality data. This will prevent multiphasic behaviour of the system by reducing steric hindrance and mass transport. However, with low ligand densities and low response levels, noise and drift become important factors (5). Equilibrate the sensor surface thoroughly and match flow and analyte buffer carefully. The noise of the system can be measured by setting a report point of 60 seconds on a stable baseline. The noise should be
< 0.3 RU on BIACORE type of systems.
The technique of signal averaging can lead to a reduction of the noise level, assuming that all data sets are identically generated (6). The identical data sets are generated by injecting each analyte concentration several times and averaging the sensorgrams to one curve for each analyte concentration. This will reduce the noise in the resulting curves, making them easier to analyse.
Ligand integrity
Immobilization with the amine coupling protocol can affect the ligand stability and activity due to the sudden pH drop and low salt conditions (7).
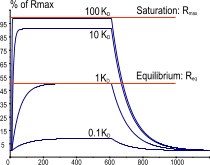
After immobilization of the ligand, the sensor surface must be checked for integrity. The best way is to inject a known binding analyte at, at least, two concentrations. For a reliable result the injection time of the highest concentration should be long enough to reach steady state. By fitting the kinetics also the Rmax is calculated. The calculated Rmax indicates how active the ligand is.
For reliable kinetics, it is not necessary to know exactly how much ligand is still biologically active. Nor is it necessary to saturate the ligand. However, knowing the actual amount of active ligand can be useful when the kinetics deviate from the 1:1 interaction. For instance, when the Rmax is lower than the expected value, consider if the active ligand sites could be blocked. When the Rmax is higher than the expected value, consider if there are more binding sites on ligand or if the analyte becomes dimerized.
Sometimes the analyte is binding promiscuously to the ligand and the sensor surface (8),(9). In the case of a promiscuous interaction, higher analyte concentrations will not saturate the surface.
In addition, the ligand integrity must be preserved even after multiple regeneration cycles. The ligand integrity can be verified after regeneration by repeatedly injecting analyte and regeneration solutions and monitoring whether the ligand binding characteristics remain comparable.
Mass transfer
Minimize mass transfer by immobilizing a small amount of ligand (see ligand density). Check for mass transfer limitation by using different flow rates. When the association and dissociation rate constants are independent of the flow rate, no mass transfer limitation is to be expected.
It becomes clear from the next table and the ‘Detecting mass transfer’ figure that for a high ligand density a flow rate of at least 40 µ/min is necessary to avoid most of the mass transport limitation. When analysing the sensorgram it is tempting to add mass transfer to the equations (10) to improve the fit. However, it is better to use the simple 1:1 model if there are no indications of mass transfer.
| High ligand | Low ligand | ||
|---|---|---|---|
| Flow rate | Injection volume | Curve slope | Curve slope |
| (µl/min) | (µl) | (RU/s) | (RU/s) |
| 5 | 10 | 4 | 9 |
| 10 | 20 | 10 | 9 |
| 20 | 40 | 17 | 10 |
| 40 | 80 | 19 | 10 |
| 80 | 160 | 20 | 11 |
References
| (1) | Myszka, D. G. Improving biosensor analysis. J.Mol.Recognit. 12: 279-284; (1999). Goto reference |
| (2) | Cheskis, B. and L. P. Freedman Modulation of nuclear receptor interactions by ligands: kinetic analysis using surface plasmon resonance. Biochemistry 35: 3309-3318; (1996). Goto reference |
| (3) | Ibrahimi, O. A., F. Zhang, S. C. Lang Hrstka, et al. Kinetic Model for FGF, FGFR, and Proteoglycan Signal Transduction Complex Assembly. Biochemistry 43: 4724-4730; (2004). Goto reference |
| (4) | Klein, P., D. Mattoon, M. A. Lemmon, et al. A structure-based model for ligand binding and dimerization of EGF receptors. Proc.Natl.Acad.Sci.U.S.A. 101: 929-934; (2004). |
| (5) | Ober, R. J. and E. S. Ward The Choice of Reference Cell in the Analysis of Kinetic Data Using BIAcore. Analytical Biochemistry 271: 70-80; (1999). |
| (6) | Ober, R. J. and E. S. Ward The influence of signal noise on the accuracy of kinetic constants measured by surface plasmon resonance experiments. Analytical Biochemistry 273: 49-59; (1999). Goto reference |
| (7) | Rich, R. L. and D. G. Myszka The Revolution of Real-Time, Label-Free Biosensor Applications. (2011). Goto reference |
| (8) | Giannetti, A. M., B. D. Koch and M. F. Browner Surface Plasmon Resonance Based Assay for the Detection and Characterization of Promiscuous Inhibitors. Journal of Medicinal Chemistry 51: 574-580; (2008). Goto reference |
| (9) | Ryan, A. J., N. M. Gray, P. N. Lowe, et al. Effect of detergent on "promiscuous" inhibitors. J.Med.Chem. 46: 3448-3451; (2003). |
| (10) | Karlsson, R. and A. Falt Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. Journal of Immunological Methods 200: 121-133; (1997). Goto reference |