Strategy
The experimental strategy is, to some extent, defined by the purpose of the experiment and the expected interaction kinetics. In principle the experimental strategy determines the amount of ligand that is immobilized on the sensor chip surface. In general, the lowest immobilization level that gives a proper interaction response should be used. The immobilization level is also dependent on the sensitivity of the instrument used.
The interaction kinetics between ligand and analyte determines the injection strategy in an experiment. In the figure 'Injection strategies', the main injection strategies are depicted.
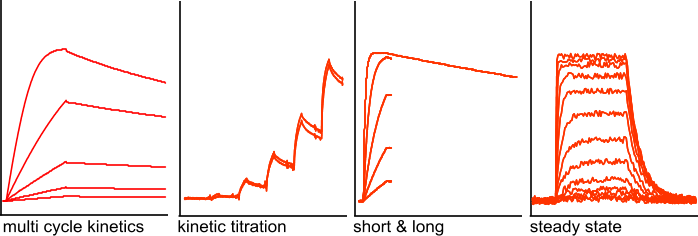
The multi cycle kinetics is the most common way to perform the experiments. Each injection of analyte or blank is done in a separate cycle. At the end of the experiment, all the single curves are put together in one sensorgram for analysis. By running single injections in an automated method, it is very easy to create an overlay of all the cycles.
The kinetic titration (single cycle kinetics) is useful with interactions that are particularly difficult to regenerate or when regeneration is detrimental to the ligand (1),(2). The analyte is injected from a low to high concentration with short dissociation times in between and a long dissociation time at the end (not shown here). All the injections are analysed in one sensorgram with a special equation for kinetic titration. The models often limit the kinetic titration to five analyte injections.
The 'short and long' experiments are done when the dissociation is very slow. Injections with low analyte concentrations take a long period to dissociate to an extent that is sufficient to be analysed. Therefore, to obtain an adequate decrease in response, only the highest analyte concentration is used for a long dissociation period. The less concentrated analyte injections are regenerated after a short dissociation period. This will greatly speed up the experiment.
Steady state experiments are possible when the dissociation rate is sufficiently fast, in general when the kd is faster than 10-3s-1. With a slow dissociation rate, it takes too long to reach steady state within one analyte injection volume (depending on the type of instrument). A steady state experiment is often used with small compounds that have a fast dissociation and therefore reach steady state quickly. With a high dissociation rate (<10-1), the association and dissociation rate are hard to determine and therefore the steady state experiment is a good alternative to get at least the equilibrium dissociation constant. In addition, a steady state experiment is frequently used to confirm the equilibrium dissociation constant.
Besides these injection schemes, there are more possibilities depending on the type of instrument. For instance, BIO-RAD offers “One-shot Kinetics™” (3),(4) with the ProteOn™ instrument. Here, one analyte is injected at several concentrations in parallel over one ligand which is immobilized on several spots (5). In the instruments with ligand arrays, such as the IBIS MX96™, one analyte at a single concentration is injected simultaneously across multiple immobilized ligand spots of different densities (6).
References
| (1) | Dougan, D. A., R. L. Malby, L. C. Gruen, et al. Effects of substitutions in the binding surface of an antibody on antigen affinity. Protein Eng 11: 65-74; (1998). Goto reference |
| (2) | Karlsson, R., P. S. Katsamba, H. Nordin, et al. Analyzing a kinetic titration series using affinity biosensors. Analytical Biochemistry 349: 136-147; (2006). Goto reference |
| (3) | Rich, R. L., G. A. Papalia, P. J. Flynn, et al. A global benchmark study using affinity-based biosensors. Analytical Biochemistry 386: 194-216; (2008). Goto reference |
| (4) | Nahshol, O., V. Bronner, A. Notcovich, et al. Parallel kinetic analysis and affinity determination of hundreds of monoclonal antibodies using the ProteOn XPR36. Analytical.Biochemistry 383: 52-60; (2008). Goto reference |
| (5) | Abdiche, Y. N., K. C. Lindquist, A. Pinkerton, et al. Expanding the ProteOn XPR36 biosensor into a 36-ligand array expedites protein interaction analysis. Analytical Biochemistry 411: 139-151; (2011). Goto reference |
| (6) | Rebe Raz, S., M. G. E. G. Bremer, M. Giesbers, et al. Development of a biosensor microarray towards food screening, using imaging surface plasmon resonance. Biosensors and Bioelectronics 24: 552-557; (2008). Goto reference |
Footnotes
- ProteOn XPR 36 is not sold any more