These are the posts of the old forum. It was not possible to transfer the user data, so they are missing in most of the posts. For new questions, go to the general discussions.
Protein-small inhibitors interaction
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
12 years 3 months ago - 12 years 2 months ago #1
by OldForum
Protein-small inhibitors interaction was created by OldForum
Hi,
initially, thank you for this fine website - I started with SPR three weeks ago and it was really greatly helpful to me.
I would like to ask you for advice concerning study of interaction of Mre11 protein with small inhibitors. I performed the analysis using GLH chip and obtained good curves and reproducible results with one of the inhibitors (plus one protein-interacting partner as a control), however I don´t know how (or if) to interpret the curves of some other ones. There was a small percentage of DMSO in the samples (0.8 %), so I performed EVC correction and used PBST buffer with the same % of DMSO - however I admit that not always as it did not seem to help much... As a double reference I used an empty activated/deactivated channel. I tried to optimize protein immobilization but it did not help and at this point I don´t know what else to possibly take into account. I will try to upload some curves if you could please take a look. I will be grateful for any suggestions.
Many thanks, Jara
initially, thank you for this fine website - I started with SPR three weeks ago and it was really greatly helpful to me.
I would like to ask you for advice concerning study of interaction of Mre11 protein with small inhibitors. I performed the analysis using GLH chip and obtained good curves and reproducible results with one of the inhibitors (plus one protein-interacting partner as a control), however I don´t know how (or if) to interpret the curves of some other ones. There was a small percentage of DMSO in the samples (0.8 %), so I performed EVC correction and used PBST buffer with the same % of DMSO - however I admit that not always as it did not seem to help much... As a double reference I used an empty activated/deactivated channel. I tried to optimize protein immobilization but it did not help and at this point I don´t know what else to possibly take into account. I will try to upload some curves if you could please take a look. I will be grateful for any suggestions.
Many thanks, Jara
Last edit: 12 years 2 months ago by OldForum.
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
12 years 3 months ago #2
by OldForum
Replied by OldForum on topic Protein-small inhibitors interaction
Hi Jara,
I am not familiar with the Bio-rad software but from your description i would say you are doing well.
In the EVC plot it looks that the binding is mass-transport limited.
However the kinetic plot look better. Is it possible to inject the analyte a little longer (e.g. 3'), so that all curves level out to steady state?
That makes it possible to do some equilibrium analysis.
In addition you should do some replicates to confirm the stability and reprocebility of the system.
What are the molecular sizes of Mre11 and the inhibitors and how much did you immobilize per spot?
regards
Arnoud
I am not familiar with the Bio-rad software but from your description i would say you are doing well.
In the EVC plot it looks that the binding is mass-transport limited.
However the kinetic plot look better. Is it possible to inject the analyte a little longer (e.g. 3'), so that all curves level out to steady state?
That makes it possible to do some equilibrium analysis.
In addition you should do some replicates to confirm the stability and reprocebility of the system.
What are the molecular sizes of Mre11 and the inhibitors and how much did you immobilize per spot?
regards
Arnoud
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
12 years 3 months ago #3
by OldForum
Replied by OldForum on topic Protein-small inhibitors interaction
Dear Arnoud,
many thanks for looking at my results.
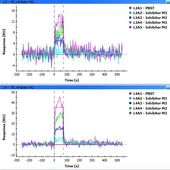
The injection time I used was at most 120 s (flow rate 80-100 ul/min) and I am prepared to try more, the problem is that the results are not reproducible - I tried a number of replications and I was never able to repeat the result as in the second picture (M2 kinetics). In fact, I obtained those better pictures after six days of using the chip so I cannot be sure what was the protein condition. In most other experiments the signal of the inhibitor seemed to be quite nonlinear (e.g. big jump between 50 uM and 100 uM concentration or sometimes even between 80 and 100 uM) and not stable (it usually does not remain constant when repeated with the same concentrations of inhibitor). It seems to me the response of the inhibitor is buried within something nonspecific...
The Mre11 protein is 77 kDa and the inhibitors are about 0.426 kDa (M2). The amount of protein immobilized was 12.5 - 60 ug/ml (that gave about 400-2000 RU).
Currently I am making no progress as I am waiting for new chips - I have some left but they are expired and I hesitate to use them.
Many thanks again,
Jara
many thanks for looking at my results.
The injection time I used was at most 120 s (flow rate 80-100 ul/min) and I am prepared to try more, the problem is that the results are not reproducible - I tried a number of replications and I was never able to repeat the result as in the second picture (M2 kinetics). In fact, I obtained those better pictures after six days of using the chip so I cannot be sure what was the protein condition. In most other experiments the signal of the inhibitor seemed to be quite nonlinear (e.g. big jump between 50 uM and 100 uM concentration or sometimes even between 80 and 100 uM) and not stable (it usually does not remain constant when repeated with the same concentrations of inhibitor). It seems to me the response of the inhibitor is buried within something nonspecific...
The Mre11 protein is 77 kDa and the inhibitors are about 0.426 kDa (M2). The amount of protein immobilized was 12.5 - 60 ug/ml (that gave about 400-2000 RU).
Currently I am making no progress as I am waiting for new chips - I have some left but they are expired and I hesitate to use them.
Many thanks again,
Jara
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
12 years 3 months ago #4
by OldForum
Replied by OldForum on topic Protein-small inhibitors interaction
Dear Jara,
Form what you write, I conclude that you have to optimize the system first.
You did immobilize a good range of Mre11. However, when I calculate the maximal response I come to
(1:1 interaction)
- 400 RU Mre11 77000 Da and M2 426 Da --> 2 RU !
- 2000 RU Mre11 77000 Da and M2 426 Da --> 11 RU
So your expected Rmax is fairly low is this combination.
Looking at your M2 kinetic figure L2 is in the expected range but I suspect that this is the 400 RU Mre11 since the second figure is higher.
It is possible that your are looking at some non-specific interaction but it looks like the overall (both specific and non-specific) interaction is not very strong.
To overcome the big jumps, you should investigate the solubility of the M2-compound (DMSO concentration).
In addition, when preparing a dilution series, make a stock from where you make a stepwise dilution in flow buffer. This will minimize buffer jumps.
It can help to start your injection series with some blank injection with analyte buffer to prime and stabilize the surface.
Some literature I can recommend:
1. Bravman, T. et al; Exploring "one-shot" kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal.Biochem. (358): 281-288; 2006.
2. Myszka, D. G.; Analysis of small-molecule interactions using Biacore S51 technology. Anal.Biochem. (329): 316-323; 2004.
3. Myszka, D. G. and Rich, R. L.; Implementing surface plasmon resonance biosensors in drug discovery. Pharm.Sci.Technol.Today. (3): 310-317; 2000.
4. Nordin, H. et al; Kinetic studies of small molecule interactions with protein kinases using biosensor technology. Anal.Biochem. (340): 359-368; 2005.
regards
Arnoud
Form what you write, I conclude that you have to optimize the system first.
You did immobilize a good range of Mre11. However, when I calculate the maximal response I come to
(1:1 interaction)
- 400 RU Mre11 77000 Da and M2 426 Da --> 2 RU !
- 2000 RU Mre11 77000 Da and M2 426 Da --> 11 RU
So your expected Rmax is fairly low is this combination.
Looking at your M2 kinetic figure L2 is in the expected range but I suspect that this is the 400 RU Mre11 since the second figure is higher.
It is possible that your are looking at some non-specific interaction but it looks like the overall (both specific and non-specific) interaction is not very strong.
To overcome the big jumps, you should investigate the solubility of the M2-compound (DMSO concentration).
In addition, when preparing a dilution series, make a stock from where you make a stepwise dilution in flow buffer. This will minimize buffer jumps.
It can help to start your injection series with some blank injection with analyte buffer to prime and stabilize the surface.
Some literature I can recommend:
1. Bravman, T. et al; Exploring "one-shot" kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal.Biochem. (358): 281-288; 2006.
2. Myszka, D. G.; Analysis of small-molecule interactions using Biacore S51 technology. Anal.Biochem. (329): 316-323; 2004.
3. Myszka, D. G. and Rich, R. L.; Implementing surface plasmon resonance biosensors in drug discovery. Pharm.Sci.Technol.Today. (3): 310-317; 2000.
4. Nordin, H. et al; Kinetic studies of small molecule interactions with protein kinases using biosensor technology. Anal.Biochem. (340): 359-368; 2005.
regards
Arnoud
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
12 years 2 months ago #5
by OldForum
Replied by OldForum on topic Protein-small inhibitors interaction
Dear Arnoud,
thanks a lot. I will follow your suggestions and check the literature recommended.
Best regards,
Jara
thanks a lot. I will follow your suggestions and check the literature recommended.
Best regards,
Jara
Please Log in or Create an account to join the conversation.