These are the posts of the old forum. It was not possible to transfer the user data, so they are missing in most of the posts. For new questions, go to the general discussions.
Signal decreasing during association phase
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 6 months ago #1
by OldForum
Signal decreasing during association phase was created by OldForum
Hi,
Could you please help with the problem I faced.
I'm using the SPR (Biacore 3000) to characterize lectin-carbohydrate interactions through kinetic and thermodynamic parameters. In our case, the sugar is immobilized onto a sensor surface (CM5 chip) through a tailor-made peptide module and lectins are flown across.
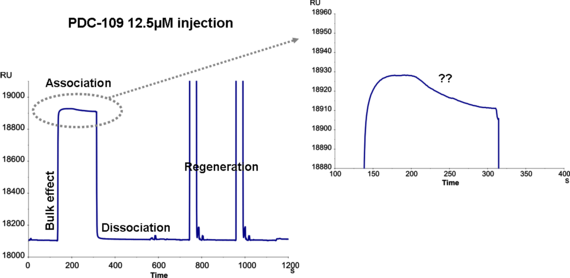
We ran a protein PDC-109 16kDa, purified from a biological sample, through a surface with the carbohydrate ligand (see file attached). I am confused with the fact that during the injection the signal in association phase is decreasing, although the analyte is still being injected. I saw the same effect in the reference surface and in different chips so I don't think that it's an artefact. Moreover, using a commercial lectin (UEA, Ulex europeaus agglutinin) or injecting running buffer in the same surfaces, normal sensorgrams are observed with no decreasing signal.
Have you had a similar experience with a signal decreasing during association phase? What may be the cause? How can I resolve this effect in order to obtain correct fittings?
I would be grateful for your reply.
Best regards,
Sira
Could you please help with the problem I faced.
I'm using the SPR (Biacore 3000) to characterize lectin-carbohydrate interactions through kinetic and thermodynamic parameters. In our case, the sugar is immobilized onto a sensor surface (CM5 chip) through a tailor-made peptide module and lectins are flown across.
We ran a protein PDC-109 16kDa, purified from a biological sample, through a surface with the carbohydrate ligand (see file attached). I am confused with the fact that during the injection the signal in association phase is decreasing, although the analyte is still being injected. I saw the same effect in the reference surface and in different chips so I don't think that it's an artefact. Moreover, using a commercial lectin (UEA, Ulex europeaus agglutinin) or injecting running buffer in the same surfaces, normal sensorgrams are observed with no decreasing signal.
Have you had a similar experience with a signal decreasing during association phase? What may be the cause? How can I resolve this effect in order to obtain correct fittings?
I would be grateful for your reply.
Best regards,
Sira
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 6 months ago - 10 years 10 months ago #2
by OldForum
Replied by OldForum on topic Signal decreasing during association phase
Hi Sira,
My first impression is that the flow buffer and your analyte do not match very wel. Is the senorgram you show before or after reference subtraction?
Because a commercial lectin does not have this problem, I think that you have some mixed affinities in your preparation.
Did you run your isolation on gel to determine the purity. Perhaps you can purify it further with HPLC.
If you have mixed affinities in your sample it is possible that a smaller molecule is binding slower to your ligand, replacing the faster binding analyte.
This will result in the shape you observed. This is a competing reaction between two analytes for the same ligand.
You can look at www.sprpages.nl/data-fitting/models.html
Regards
Arnoud
My first impression is that the flow buffer and your analyte do not match very wel. Is the senorgram you show before or after reference subtraction?
Because a commercial lectin does not have this problem, I think that you have some mixed affinities in your preparation.
Did you run your isolation on gel to determine the purity. Perhaps you can purify it further with HPLC.
If you have mixed affinities in your sample it is possible that a smaller molecule is binding slower to your ligand, replacing the faster binding analyte.
This will result in the shape you observed. This is a competing reaction between two analytes for the same ligand.
You can look at www.sprpages.nl/data-fitting/models.html
Regards
Arnoud
Last edit: 10 years 10 months ago by .
Please Log in or Create an account to join the conversation.