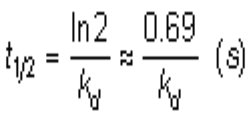These are the posts of the old forum. It was not possible to transfer the user data, so they are missing in most of the posts. For new questions, go to the general discussions.
Dissociation phase and injection time issues
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago #1
by OldForum
Dissociation phase and injection time issues was created by OldForum
Hi all,
I'm just starting out with the Biacore T200, and I've run into some problems I don't understand. We also have several contacts at the company, who keep saying different things, adding to the confusion. So, I have 2 main questions:
1) In our lab, we're producing different antibodies against c-reactive protein (CRP). We're using the SPR to do kinetics analysis to compare them on the basis of affinity. At first, we were using the CM5 chip, and we were aiming for immobilization levels of about 100 RU, and the fittings worked OK. However, we later found out that our dissociation phase was apparently not what it was supposed to be, because it resembled a near horizontal line. No decrease to the baseline. They told us that this was due to the fact that CRP is a pentamere, and so there were probably still too many multivalent interactions going on. Two ways to solve this, they said, was aim for much lower RU's, like 10 RU, and switch to the CM1 chip, because that's only a 2-dimensional dextran layer. So we did both, as they suggested. But aiming for 10 RU doesn't work. It keeps saying "response too slow". So we then simply picked a concentration, and we obtained an RU of 18 when we substracted the EDC/NHS level from the immobilization level. First thing: what does the machine mean when it "aims for 10 RU"? Is it the total RU after EDC, NHS and antibody? Because then it's impossible to ever get to 10 RU, because the EDC/NHS already reaches around 50 RU. Or is it the actual antibody effect (so immobilization level - EDC/NHS level)? In that case, 18 RU would be in the neighbourhood of what they advised. And secondly: now the fittings don't work anymore and the dissociation is still a near flat horizontal line and no decrease. What are we doing wrong? Is the amount of antibody too low now for the machine to be able to do anything with the curves? Because you do see nicely different curves when you bind with different concentrations of CRP. Albeit they are al little bit noisy. We used the same CRP concentrations as with the CM5 chips with the higher immobilization levels. Were those too high? Do we need to do a new pH scouting? And how come the dissociation needs to return to the baseline? I'm being told that the only thing that's actually happening is that you switch back to the running buffer (so you're basically washing away the excess), at the same flow rate. Can that actually dissociate the antigens from the antibodies?
2) In another application field, we're using the SA chips to immobilize phages expressing biotin. When we do startup cycles using PBS and we specify the injection time to 300 seconds, either on a naked SA chip or on an immobilized SA chip, the response of the PBS shoots down to several thousands of RU, stays there for about 70 seconds, and then returns to the baseline. When we add sample, it shoots even further down than the pure PBS, stays there for about 150 seconds, and then also returns to baseline. Why is the specified contact time ignored? It's like the flow suddenly stops or something, and we thought about a blockage of some sorts, but in the next startup cycle, it just does the exact same thing.
Can anybody give me some useful tips? Thanks a bunch!
Haley
I'm just starting out with the Biacore T200, and I've run into some problems I don't understand. We also have several contacts at the company, who keep saying different things, adding to the confusion. So, I have 2 main questions:
1) In our lab, we're producing different antibodies against c-reactive protein (CRP). We're using the SPR to do kinetics analysis to compare them on the basis of affinity. At first, we were using the CM5 chip, and we were aiming for immobilization levels of about 100 RU, and the fittings worked OK. However, we later found out that our dissociation phase was apparently not what it was supposed to be, because it resembled a near horizontal line. No decrease to the baseline. They told us that this was due to the fact that CRP is a pentamere, and so there were probably still too many multivalent interactions going on. Two ways to solve this, they said, was aim for much lower RU's, like 10 RU, and switch to the CM1 chip, because that's only a 2-dimensional dextran layer. So we did both, as they suggested. But aiming for 10 RU doesn't work. It keeps saying "response too slow". So we then simply picked a concentration, and we obtained an RU of 18 when we substracted the EDC/NHS level from the immobilization level. First thing: what does the machine mean when it "aims for 10 RU"? Is it the total RU after EDC, NHS and antibody? Because then it's impossible to ever get to 10 RU, because the EDC/NHS already reaches around 50 RU. Or is it the actual antibody effect (so immobilization level - EDC/NHS level)? In that case, 18 RU would be in the neighbourhood of what they advised. And secondly: now the fittings don't work anymore and the dissociation is still a near flat horizontal line and no decrease. What are we doing wrong? Is the amount of antibody too low now for the machine to be able to do anything with the curves? Because you do see nicely different curves when you bind with different concentrations of CRP. Albeit they are al little bit noisy. We used the same CRP concentrations as with the CM5 chips with the higher immobilization levels. Were those too high? Do we need to do a new pH scouting? And how come the dissociation needs to return to the baseline? I'm being told that the only thing that's actually happening is that you switch back to the running buffer (so you're basically washing away the excess), at the same flow rate. Can that actually dissociate the antigens from the antibodies?
2) In another application field, we're using the SA chips to immobilize phages expressing biotin. When we do startup cycles using PBS and we specify the injection time to 300 seconds, either on a naked SA chip or on an immobilized SA chip, the response of the PBS shoots down to several thousands of RU, stays there for about 70 seconds, and then returns to the baseline. When we add sample, it shoots even further down than the pure PBS, stays there for about 150 seconds, and then also returns to baseline. Why is the specified contact time ignored? It's like the flow suddenly stops or something, and we thought about a blockage of some sorts, but in the next startup cycle, it just does the exact same thing.
Can anybody give me some useful tips? Thanks a bunch!
Haley
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago #2
by OldForum
Replied by OldForum on topic Dissociation phase and injection time issues
1.1. Complex half life time
The complex half-life time is the time needed for dissociation of half of the complexes to their individual components (1).
A strong binding (> 10-4 s-1) is difficult to analyse when the dissociation curve is to short. As a rule of thumb, the dissociation curve should decrease at least 5% before analysis is attempted. For a dissociation constant of 10-4 s-1 this will result in a dissociation time of at least 12 minutes. In addition, sufficient blank injections with the same long "dissociation" times are needed to compensate for possible baseline drift (5).
Long dissociation times are not practical when done with every injection. Instead, only several long dissociations are necessary, and the rest of the experiment can have short dissociation times (200–300 s). Use the long dissociations for the highest injected analyte concentrations (5). However, it is important to avoid analyte concentration that can introduce artefacts such as non-specific binding, steric hindrance or aggregation.
1.2. In practise
From your question, I deduce that you immobilize the antibodies (150000 Da). The analyte is a pentameter (CRP; 5x 25106 Da (125530 Da) but the antibodies are dimers. It may be better to immobilize the CRP, use the antibodies as analyte, and thus lower the rebinding change. The use of C1 can also help lower the rebinding because is has no dextran layer and alone carboxyl groups lowering the amount of bound ligand.
As you tried to lower the amount of bound ligand to 10 RU. The expected Rmax of the system is thus around 8 RU (10 x (125530/150000). As you not include a sensorgram or Rmax I’am not sure in what range your response is. I know that this kind of response are possible with a T200 but I would not recommend this with this low dissociation rates.
I’am not familiar with the way of how the T200 software is aiming for the immobilization level. Normally, I look at the difference between then response after activation and the response after deactivation. This is a rough estimation of how much ligand is bound. The real amount is not important as long as the kinetics is all right (sufficient response, association curvature, enough dissociation). I think the ‘response to slow’ means that the amount of ligand pre-concentration is to slow. As you increase the ligand concentration this will resolve.
As you can calculate from equation the complex half time is dependent on the kd. In case of a flat line the kd is lower than 10-5 s-1; which will give a half life of at least 19 hours! For a proper kd-calculation the dissociation must be at least 5%. Thus when the dissociation starts at 100 RU the curve should at least lower to 95 RU before fitting is attempted. You can imaging when the response is very low this will be difficult to achieve because 5% of 10 RU is 0.5 RU and that will be probably within the noise. A second problem is the long dissociation time during which the system possibly will have some drift. Having proper double referenced curves is a must is this case.
Some steps you can take:
· Immobilize CRP or antibodies (you can make a low (10 – 20 RU) and high channel (50 – 100 RU)
· Create an appropriate reference channel
· Equilibrate overnight the chip
· Inject flow buffer only as a blank (3x !)
· Inject CRP or antibodies
· end with flow buffer injection as blank control
Best way to do this in an automated method with sufficient equilibrium time before injection and long dissociation times.
The complex half-life time is the time needed for dissociation of half of the complexes to their individual components (1).
A strong binding (> 10-4 s-1) is difficult to analyse when the dissociation curve is to short. As a rule of thumb, the dissociation curve should decrease at least 5% before analysis is attempted. For a dissociation constant of 10-4 s-1 this will result in a dissociation time of at least 12 minutes. In addition, sufficient blank injections with the same long "dissociation" times are needed to compensate for possible baseline drift (5).
Long dissociation times are not practical when done with every injection. Instead, only several long dissociations are necessary, and the rest of the experiment can have short dissociation times (200–300 s). Use the long dissociations for the highest injected analyte concentrations (5). However, it is important to avoid analyte concentration that can introduce artefacts such as non-specific binding, steric hindrance or aggregation.
1.2. In practise
From your question, I deduce that you immobilize the antibodies (150000 Da). The analyte is a pentameter (CRP; 5x 25106 Da (125530 Da) but the antibodies are dimers. It may be better to immobilize the CRP, use the antibodies as analyte, and thus lower the rebinding change. The use of C1 can also help lower the rebinding because is has no dextran layer and alone carboxyl groups lowering the amount of bound ligand.
As you tried to lower the amount of bound ligand to 10 RU. The expected Rmax of the system is thus around 8 RU (10 x (125530/150000). As you not include a sensorgram or Rmax I’am not sure in what range your response is. I know that this kind of response are possible with a T200 but I would not recommend this with this low dissociation rates.
I’am not familiar with the way of how the T200 software is aiming for the immobilization level. Normally, I look at the difference between then response after activation and the response after deactivation. This is a rough estimation of how much ligand is bound. The real amount is not important as long as the kinetics is all right (sufficient response, association curvature, enough dissociation). I think the ‘response to slow’ means that the amount of ligand pre-concentration is to slow. As you increase the ligand concentration this will resolve.
As you can calculate from equation the complex half time is dependent on the kd. In case of a flat line the kd is lower than 10-5 s-1; which will give a half life of at least 19 hours! For a proper kd-calculation the dissociation must be at least 5%. Thus when the dissociation starts at 100 RU the curve should at least lower to 95 RU before fitting is attempted. You can imaging when the response is very low this will be difficult to achieve because 5% of 10 RU is 0.5 RU and that will be probably within the noise. A second problem is the long dissociation time during which the system possibly will have some drift. Having proper double referenced curves is a must is this case.
Some steps you can take:
· Immobilize CRP or antibodies (you can make a low (10 – 20 RU) and high channel (50 – 100 RU)
· Create an appropriate reference channel
· Equilibrate overnight the chip
· Inject flow buffer only as a blank (3x !)
· Inject CRP or antibodies
· end with flow buffer injection as blank control
Best way to do this in an automated method with sufficient equilibrium time before injection and long dissociation times.
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago #3
by OldForum
Replied by OldForum on topic Dissociation phase and injection time issues
2) Your second part look like if there is something wrong with the system or sensor chip. These kind of jumps are not normal. When you inject the flow buffer, every curve should be within plus or minus 5 RU. You can inject for instance flow buffer with added 10 mM NaCl, which should give an positive injection buffer jump during the injection. If this is not the case check the system with a maintenance or new sensor chip.
Please Log in or Create an account to join the conversation.